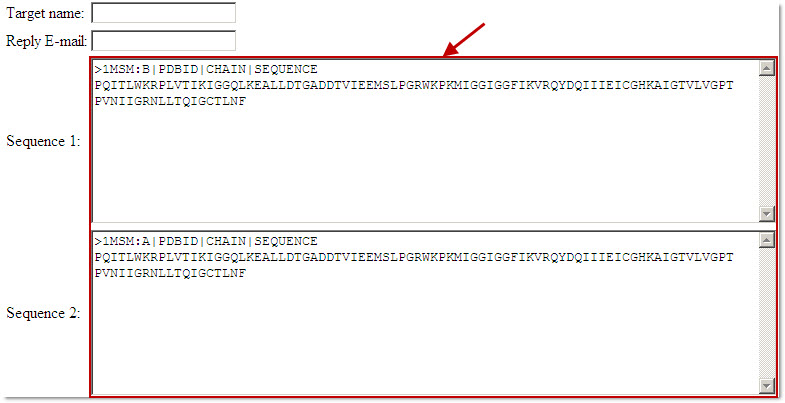
Proinfo PPC server requires the user to input the target amino acid sequences by copying and pasting "each sequence" into a separate input box. The user can either input the sequence in FASTA format or the amino acid sequence without the sequence identifier.

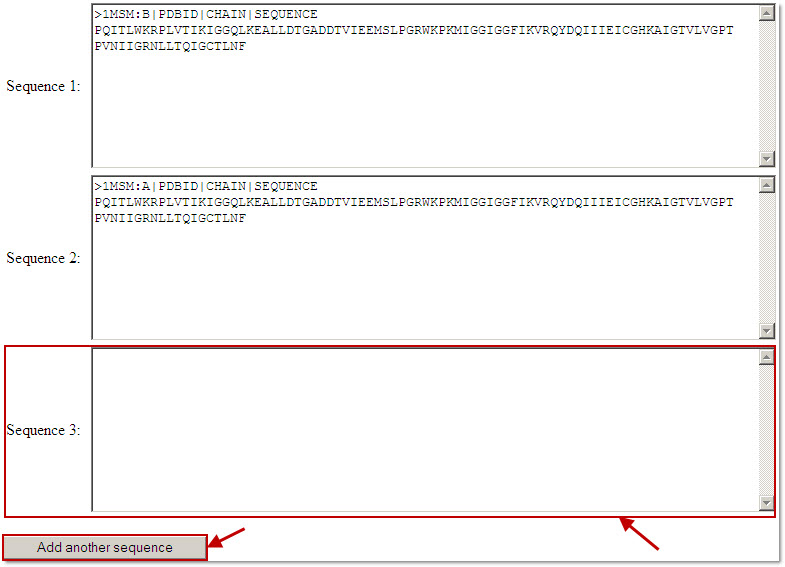
Users can add more amino acid sequences by clicking the 'add another sequence' button below the input box.
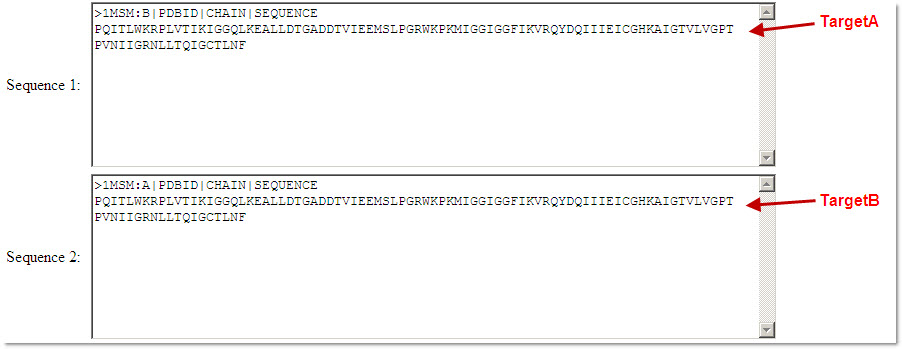
The web server automatically assigns a unique ID for each amino acid sequence, starting from TargetA, according to the sequence order submitted by the user. Please note that sequence identifiers that were submitted with the sequences (in FASTA format) will be obmitted.
A name has to be provided with the submitted job and it is also required that user provide an email address to where the resulting protein complex models will be sent.
Please note that some email service providers appear to reformat email messages, which in turn breaks the PDB format of results returned by Protinfo PPC. The following list shows some of the emails that may not display Proinfo PPC result correctly.
Since modeling a large protein complex that consists of several protein chains usually takes a considerable amount of time, the maximum number of input amino sequences is limited to five.
In addition, users can instruct Protinfo PPC server to mark interacting residues in the output PDB files by selecting "Mark interacting residues usign B-factor column" box. The value of B-factor column will be 1 if the residue is interacting with other residues and 0 otherwise. We defined interacting residues to be those that have at least one atom that is closer than 5 angstrom to another atom of another residue from a different polypeptide chain.
Lastly, Protinfo PPC server provides users with the option to receive the coordinates of the corresponding initial models that were used to create the final complex models. This is essentially PDB model without the coordinates of the residues that are within loop regions (insertion/deletion regions).
Protinfo PPC server returns the resulting protein complex models in a standard PDB CASP format to user specified email address. For each submitted job, a maximum of 5 complex models will be returned.
Remarks describing normalized RAPDF score, percentage of sequence identity between template and target and the template-target map are provided for each complex model.
A template-target map: 1LDT-T_TargetB==1LDT-L_TargetA means that PDB structure 1LDT was used as a template to model the interaction between TargetA and TargetB sequence where chain 1LDT-T has high sequence similarity to TargetB and 1LDT-L has sequence similarity to TargetA.
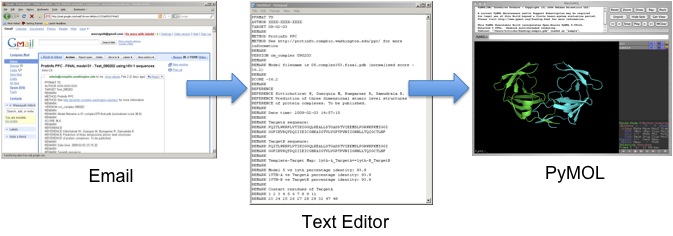
The predicted protein complex model in PDB format returned to the user's email can easily be copied and saved to a file using any text editing program. The saved file can then be visualized by using any molecular visualization softwares, such as PyMOL and CHIMERA.
The Protinfo PPC server uses existing protein complex structures to generate 3D protein complex models of the submitted sequences. If the sequences do not share any significant similarity to any of the template, no prediction will be made. Another word, there is no protein complex structure to support the interaction of the given protein sequences.
However it is possible that some of the known protein complex are missing from our protein complex library. When the server did not return any predicted model, users are encouraged to visit the Protein Data Bank (PDB) and double check to make sure that there is really no solved protein complex structure that is homologous to your sequences of interest. We usually update our protein complex template library regularly. However, some technical problems can prevent us from having the most up-to-date collection of protein complex templates. In the case where a homologous template is found in the PDB website, the user can download the PDB file and use it to model their protein sequences of interest using the custom template feature of the Protinfo PPC server.